

The only equipment needed to perform MLPA is a capillary electrophoresis device, and a thermocycler. This size range provides optimal fragment separation and low background on sequence type gels. The length increases in a stepwise-fashion, with the total size range lying between 120-500 nucleotides. An MLPA probe set is designed so that the length of each of its amplification products is unique. The number of probe ligation products of one probe therefore depends on the number of target sequences in the sample.įollowing the PCR, the resulting amplification products are separated by capillary electrophoresis.
These ligated probes are amplified exponentially during the PCR reaction, while the individual non-ligated probe oligonucleotides are not. Only when the two probe oligonucleotides are hybridised to their target can they be ligated into a single probe, containing both the forward and reverse primer sequence. MLPA: Designs probes for copy number detection and SNP studies See also: words rhyming with alleleid.
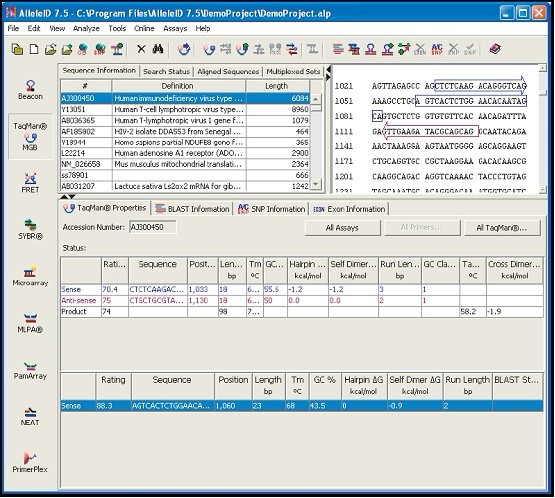
#ALLELEID MLPA SOFTWARE#
The software is designed to run on Windows and Macintosh operating system. This aids in bacterial identification, pathogen detection and species identification. These two probe oligonucleotides hybridise to immediately adjacent target sites. AlleleID designs oligos for strain differentiation and taxa specific assays.
#ALLELEID MLPA PLUS#
Each MLPA probe consists of two paired oligonucleotides, each one containing one of the PCR primer sequences plus a sequence complementary to the DNA target sequence. The trick of MLPA is that it is not the sample DNA that is amplified, but the MLPA probes that are added to the sample. In contrast, all fragments in an MLPA reaction are amplified using the same PCR primer pair, making the reaction extremely robust. For this reason, the in-silico operations including utilization of the NCBI RefSeq database, Servers of PanSeq and Gview, AlleleID 7.7 and Oligo Analyzer 3.1 was done.

Furthermore, small differences in reaction conditions can result in large differences in the obtained results as each primer pair may react slightly differently to the change. The primer efficiency of different primer pairs can vary, thus making it difficult to use conventional multiplex PCR for relative quantification of target sequences. the human BRCA1, MSH2 and MLH1 genes, detection of trisomies such as Down syndrome, characterisation of chromosomal aberrations in cell lines and tumour samples, SNP and mutation detection, and DNA methylation analysis.Ĭonventional multiplex PCR uses one pair of primers for each fragment, resulting in a reaction that contains a large number of different primer sets. Applications include: the detection of exon deletions/duplications in e.g. MLPA reactions require only 50 ng of human chromosomal DNA. © 2012 Wiley Periodicals, Inc.Multiplex Ligation-dependent Probe Amplification (MLPA ®) is a method that detects aberrant copy numbers in up to 60 specific nucleic acid sequences by performing one simple PCR reaction, using a single PCR primer pair. Our findings indicate that a combination of an open chromatin conformation and short non‐B DNA‐forming repeats may predispose to recurrent mitotic NAHR events between SUZ12 and its pseudogene.
This hotspot harbored 20% (8/40) of the type‐2 deletion breakpoints and contains the 253‐bp recurrent breakpoint region BR6 in which four independent type‐2 deletion breakpoints were identified. Further, the analysis of this large group of type‐2 deletions revealed breakpoint recurrence within short segments (ranging in size from 57 to 253‐bp) as well as the existence of a novel NAHR hotspot of 1.9‐kb (termed PRS4). Breakpoint analysis of these 23 type‐2 deletions, together with 17 NAHR‐mediated type‐2 deletions identified previously, revealed that the breakpoints are nonuniformly distributed within the paralogous SUZ12 and SUZ12P sequences. Here, a custom‐designed MLPA‐ and PCR‐based approach was used to identify 23 novel NAHR‐mediated type‐2 NF1 deletions. Type‐2 NF1 microdeletions, which are predominantly of postzygotic origin, constitute a highly informative model with which to investigate the features of mitotic NAHR. Although several disease‐associated meiotic NAHR breakpoints have been analyzed in great detail, hotspots for mitotic NAHR are not well characterized. Nonallelic homologous recombination (NAHR) is one of the major mechanisms underlying copy number variation in the human genome. Messiaen, Ludwine Kehrer‐Sawatzki, Hildegard Vogt, Julia Mussotter, Tanja Bengesser, Kathrin Claes, Kathleen Högel, Josef Chuzhanova, Nadia Fu, Chuanhua van den Ende, Jenneke Mautner, Victor‐Felix Cooper, David N. Identification of recurrent type‐2 NF1 microdeletions reveals a mitotic nonallelic homologous recombination hotspot underlying a human genomic disorder Identification of recurrent type‐2 NF1 microdeletions reveals a mitotic nonallelic homologous.


 0 kommentar(er)
0 kommentar(er)
